aduron
October 26, 2021, 4:46pm
1
I'm going through the tutorial at Tutorial: Build a Knowledge Graph using NLP and Ontologies - Developer Guides
Once I get to slide 15 I'm getting a no changes, no records. Possibly something to do with HAS_ENTITY? Any help appreciated thanks:
MATCH (c:Category {name: "NoSQL database management system"})
neo4j desktop version 1.4.8
which plugins / extensions / procedures do you use: APOC, n10s
Hello
Please install GDS plugin that is compatible with desktop version that you have installed.
Thanking you
Also install neosemantics 10s plugin
Thanking you
DMF194
February 25, 2022, 8:48am
4
Hi there,
I have also started following at Tutorial: Build a Knowledge Graph using NLP and Ontologies - Developer Guides .
Able to following along with no issues until the part where I need to import RDF from WikiData
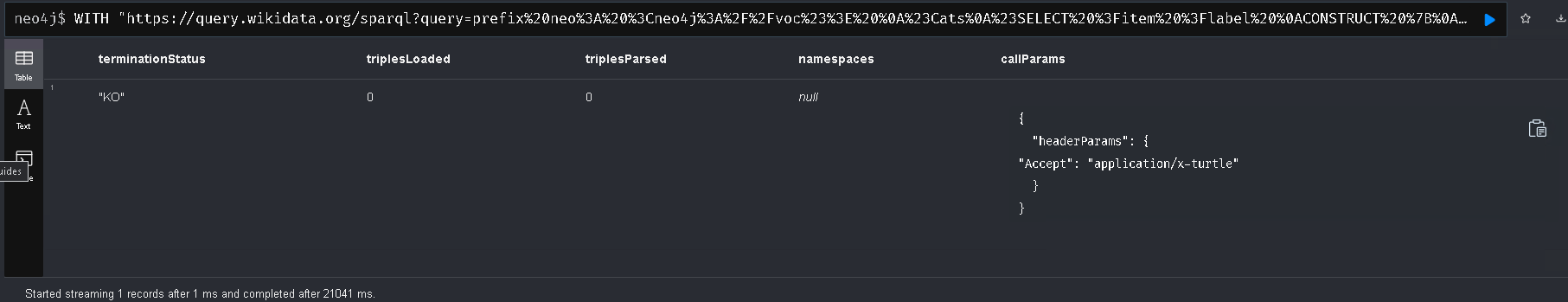
WITH "https://query.wikidata.org/sparql?query=prefix%20neo%3A%20%3Cneo4j%3A%2F%2Fvoc%23%3E%20%0A%23Cats%0A%23SELECT%20%3Fitem%20%3Flabel%20%0ACONSTRUCT%20%7B%0A%3Fitem%20a%20neo%3ACategory%20%3B%20neo%3AsubCatOf%20%3FparentItem%20.%20%20%0A%20%20%3Fitem%20neo%3Aname%20%3Flabel%20.%0A%20%20%3FparentItem%20a%20neo%3ACategory%3B%20neo%3Aname%20%3FparentLabel%20.%0A%20%20%3Farticle%20a%20neo%3AWikipediaPage%3B%20neo%3Aabout%20%3Fitem%20%3B%0A%20%20%20%20%20%20%20%20%20%20%20%0A%7D%0AWHERE%20%0A%7B%0A%20%20%3Fitem%20(wdt%3AP31%7Cwdt%3AP279)*%20wd%3AQ2429814%20.%0A%20%20%3Fitem%20wdt%3AP31%7Cwdt%3AP279%20%3FparentItem%20.%0A%20%20%3Fitem%20rdfs%3Alabel%20%3Flabel%20.%0A%20%20filter(lang(%3Flabel)%20%3D%20%22en%22)%0A%20%20%3FparentItem%20rdfs%3Alabel%20%3FparentLabel%20.%0A%20%20filter(lang(%3FparentLabel)%20%3D%20%22en%22)%0A%20%20%0A%20%20OPTIONAL%20%7B%0A%20%20%20%20%20%20%3Farticle%20schema%3Aabout%20%3Fitem%20%3B%0A%20%20%20%20%20%20%20%20%20%20%20%20schema%3AinLanguage%20%22en%22%20%3B%0A%20%20%20%20%20%20%20%20%20%20%20%20schema%3AisPartOf%20%3Chttps%3A%2F%2Fen.wikipedia.org%2F%3E%20.%0A%20%20%20%20%7D%0A%20%20%0A%7D" AS softwareSystemsUri
CALL n10s.rdf.import.fetch(softwareSystemsUri, 'Turtle' , { headerParams: { Accept: "application/x-turtle" } })
YIELD terminationStatus, triplesLoaded, triplesParsed, namespaces, callParams
RETURN terminationStatus, triplesLoaded, triplesParsed, namespaces, callParams;
For whatever reason, the result is KO
I have also added the line below in my neo4j.config file
dbms.unmanaged_extension_classes=n10s.endpoint=/rdf
OS: Windows10
Any suggestions would be advised.
Hello
As a remote observer I noticed that you are using SparkQL.Have you imported necessary ontologies for that? I guess there is a separate ontology for with name like rdf triples.Its just a guess but anyways you are the best judge for your Neo4j desktop.
Many thanks
1 Like
DMF194
February 28, 2022, 1:46am
6
HI @sameer.gijare14 ,
Yes I am using SparkQL from WikiData
Based on my current understanding, the code below is used to import taxonomies from Software Systems using the WikiData API
WITH "https://query.wikidata.org/sparql?query=prefix%20neo%3A%20%3Cneo4j%3A%2F%2Fvoc%23%3E%20%0A%23Cats%0A%23SELECT%20%3Fitem%20%3Flabel%20%0ACONSTRUCT%20%7B%0A%3Fitem%20a%20neo%3ACategory%20%3B%20neo%3AsubCatOf%20%3FparentItem%20.%20%20%0A%20%20%3Fitem%20neo%3Aname%20%3Flabel%20.%0A%20%20%3FparentItem%20a%20neo%3ACategory%3B%20neo%3Aname%20%3FparentLabel%20.%0A%20%20%3Farticle%20a%20neo%3AWikipediaPage%3B%20neo%3Aabout%20%3Fitem%20%3B%0A%20%20%20%20%20%20%20%20%20%20%20%0A%7D%0AWHERE%20%0A%7B%0A%20%20%3Fitem%20(wdt%3AP31%7Cwdt%3AP279)*%20wd%3AQ2429814%20.%0A%20%20%3Fitem%20wdt%3AP31%7Cwdt%3AP279%20%3FparentItem%20.%0A%20%20%3Fitem%20rdfs%3Alabel%20%3Flabel%20.%0A%20%20filter(lang(%3Flabel)%20%3D%20%22en%22)%0A%20%20%3FparentItem%20rdfs%3Alabel%20%3FparentLabel%20.%0A%20%20filter(lang(%3FparentLabel)%20%3D%20%22en%22)%0A%20%20%0A%20%20OPTIONAL%20%7B%0A%20%20%20%20%20%20%3Farticle%20schema%3Aabout%20%3Fitem%20%3B%0A%20%20%20%20%20%20%20%20%20%20%20%20schema%3AinLanguage%20%22en%22%20%3B%0A%20%20%20%20%20%20%20%20%20%20%20%20schema%3AisPartOf%20%3Chttps%3A%2F%2Fen.wikipedia.org%2F%3E%20.%0A%20%20%20%20%7D%0A%20%20%0A%7D" AS softwareSystemsUri
CALL n10s.rdf.import.fetch(softwareSystemsUri, 'Turtle' , { headerParams: { Accept: "application/x-turtle" } })
YIELD terminationStatus, triplesLoaded, triplesParsed, namespaces, callParams
RETURN terminationStatus, triplesLoaded, triplesParsed, namespaces, callParams;
As you said I am using the Neo4j Desktop and I am using the Neosemantics plugin in Neo4j Desktop to import the taxonomy
Is there an updated document on this as I feel it does not provide a complete picture?